
Investigation of bacterial composition by culturing techniques is laborious and prone to strong bias, since the growth requirements of many bacteria are still unknown and therefore cannot be provided under laboratory conditions. To overcome these drawbacks, culture-independent molecular methods based on amplification of 16S rRNA genes have been introduced to obtain a better understanding of the intestinal microbiota.
Regardless of the technique used for the actual DNA-based analysis, the first step is always DNA extraction and purification. Therefore, an efficient technique for the recovery of bacterial DNA is fundamental to successful molecular analysis of microbiota. If the isolated DNA does not accurately represent the microbial composition of the original sample due to inefficient cell disruption, or if the DNA purification procedure is inadequate, the any subsequent DNA-based analysis of microbial community will provide biased results. Unfortunately, the literature is rich in studies where this aspect has not been considered and the most simple and convenient DNA extraction protocol/kit has been chosen.
At Alimetrics, we can use any DNA isolation protocol requested by our client. However, we strongly urge the clients to consider the overall benefits. Samples are often unique, originating from an expensive clinical study or animal trial, and the DNA isolated from the samples may be subjected to relatively costly further analyses such as DNA sequencing. If the DNA extracted does not represent the bacterial population present in samples, the study may not fulfill its original objectives and answer the questions formulated.
Figure below illustrates the results of a comparative study in which the standard Alimetrics DNA extraction protocol outperformed the widely used commercial DNA extraction kits in the analysis of total bacterial numbers by qPCR in human faecal samples. The outcome of this molecular technique depends entirely on the total DNA yield and purity; and therefore, accurately summarises the evaluation study.
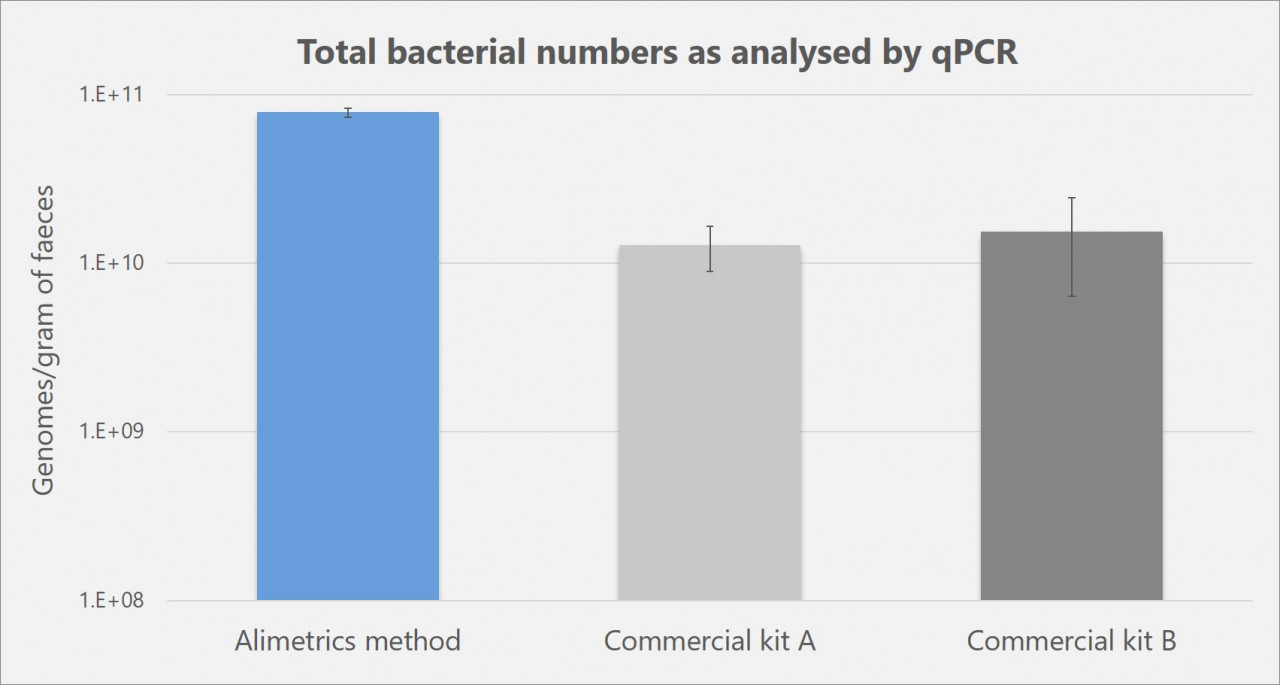
Comparison of analytical results from a human faecal specimen divided into aliquots and with replicate aliquots subjected to different DNA extraction methods. The results from the three best protocols tested are shown. Total number of bacterial 16S rRNA gene operons was determined by quantitative real-time PCR.


